treefile
select-file: Click this button to upload a tree file in plain text files, and in one of supported formats format (Newick, Nexus, phyloXML).
Example trees in Newick format:
- A tree with bootstraps and branch lengths:
(A:0.1,(B:0.1,C:0.1)90:0.1)98:0.3);
A, B, C : leaf names, also used as node IDs
0.1, 0.3 : branch lengths
90,98 : bootstrap values
- A tree with internal node IDs:
(A:0.1,(B:0.1,C:0.1)INT1:0.1[90])INT2:0.3[98]);
A, B, C : leaf names, also used as node IDs
INT1, INT2 : internal node IDs
0.1, 0.3 : branch lengths
90,98 : bootstrap values
Example trees in Nexus format:
#NEXUS
BEGIN TAXA;
DIMENSIONS NTAX = 5;
TAXLABELS
t2
t1
t5
t4
t3
;
END;
BEGIN TREES;
TRANSLATE
1 t2,
2 t1,
3 t5,
4 t4,
5 t3
;
TREE one = [&R]
((1:0.04,2:0.34):0.89,(3:0.37,(4:0.03,5:0.67):0.9):0.59);
TREE two = [&U] (1,3,(5,(2,4));
END;
- The
TRANSLATEcommand provides short alternatives to the taxon names, making the tree descriptions shorter (takes up fewer bytes of disk space). - the translate command is not necessary however; it is ok to use the taxon names directly in the tree descriptions
- the
TREEcommand denotes the start of a tree description, which consists of a tree name (e.g.,oneandtwoare used here), followed by an equals sign and then the tree topology in the standard, parenthetical notation (often referred to as the Newick or New Hampshire format). - The special comments consisting of an ampersand symbol followed by the letter U tell PAUP* to interpret the tree as being an unrooted tree.
- Files containing only the
#NEXUSplus a trees block are called tree files.
#NEXUS
BEGIN TREES;
UTREE 1 = (((Carp: 2.951519, Fugu: 2.951519) [&95%={1.904, 3.822}]: 1.237256, (Frog: 3.410711, Human: 3.410711) [&95%={3.303, 3.502}]: 0.778064) [&95%={4.160, 4.220}]: 0.251817, (Shark: 3.247244, Stingray: 3.247244) [&95%={2.258, 4.129}]: 1.193348) [&95%={4.225, 4.631}];
END;
Carp, Fugu,... : leaf names, also used as node IDs
[&95%={2.258, 4.129}] : confidence interval of divergence time
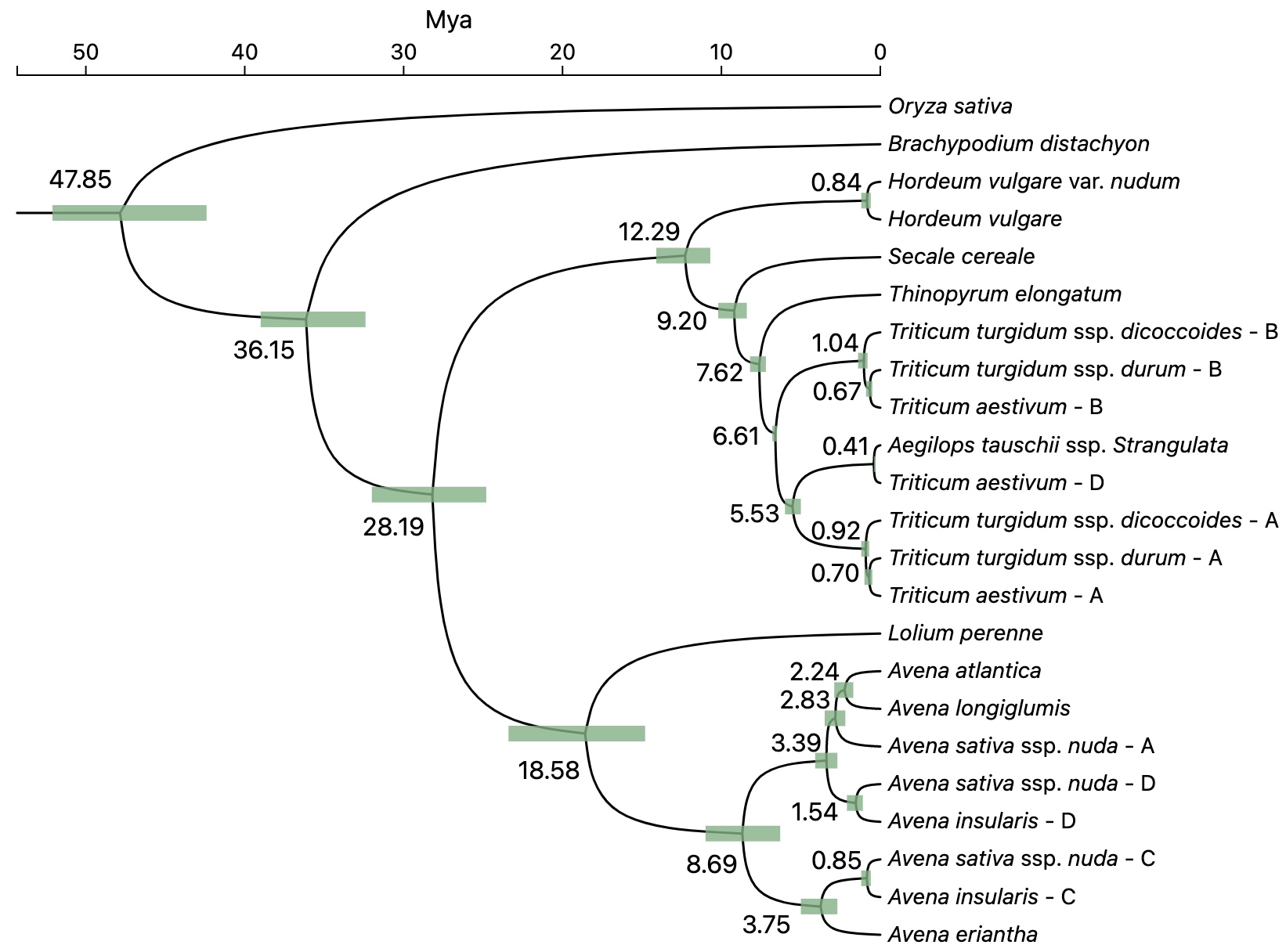
Example divergence time tree: View Tree
view: Click this button to view the tree file, only valid after uploading a tree file.
copy-leavesID: Copy all node IDs of the tree leaves to the system clipboard for easy inclusion in annotation data files.
We strongly recommend that users use simple IDs to build phylogenetic trees.
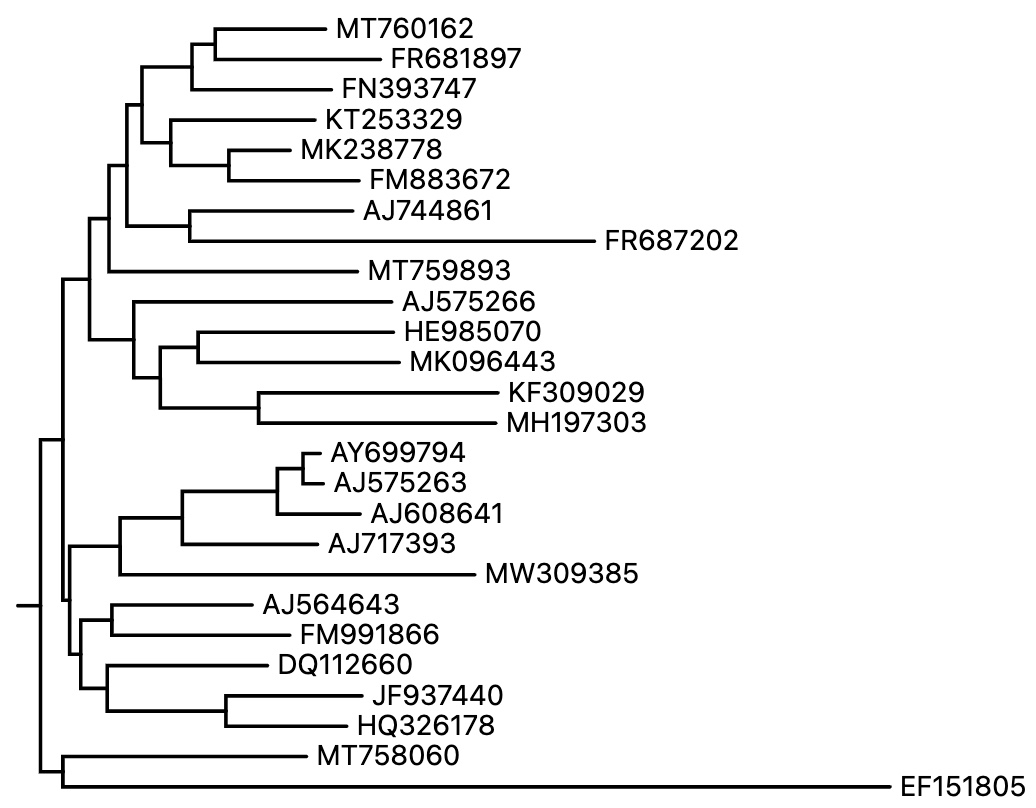
The tree above will generate the below IDs:
MT760162
FR681897
FN393747
KT253329
MK238778
FM883672
AJ744861
FR687202
MT759893
AJ575266
HE985070
MK096443
KF309029
MH197303
AY699794
AJ575263
AJ608641
AJ717393
MW309385
AJ564643
FM991866
DQ112660
JF937440
HQ326178
MT758060
EF151805
copy-internalID: Copy all IDs of internal nodes to the system clipboard for easy inclusion in annotation data files. Internal nodes IDs will be used in some annotation datasets which add external information to the tree branches, such as text labels and pie chart.
Tree file already has internal node IDs present (as shown in the second example tree in Newick format above): In this case, you can use these IDs directly in all annotation data files.
There are no internal node IDs in tree file : A unique ID will be generated for each internal node.
Copy ID of this node function to copy each Internal node ID.
Restore drawing state
We use file in json format to save the final drawing state of svg canvas, which is described in more detail in tree export chapter.
select-file: Click this button to upload a json file, and the final drawing state of svg canvas will be restored for easy secondary modification.
Example data
view: Click this button to view the example tree file used in the page opened for the first time.